Documents index
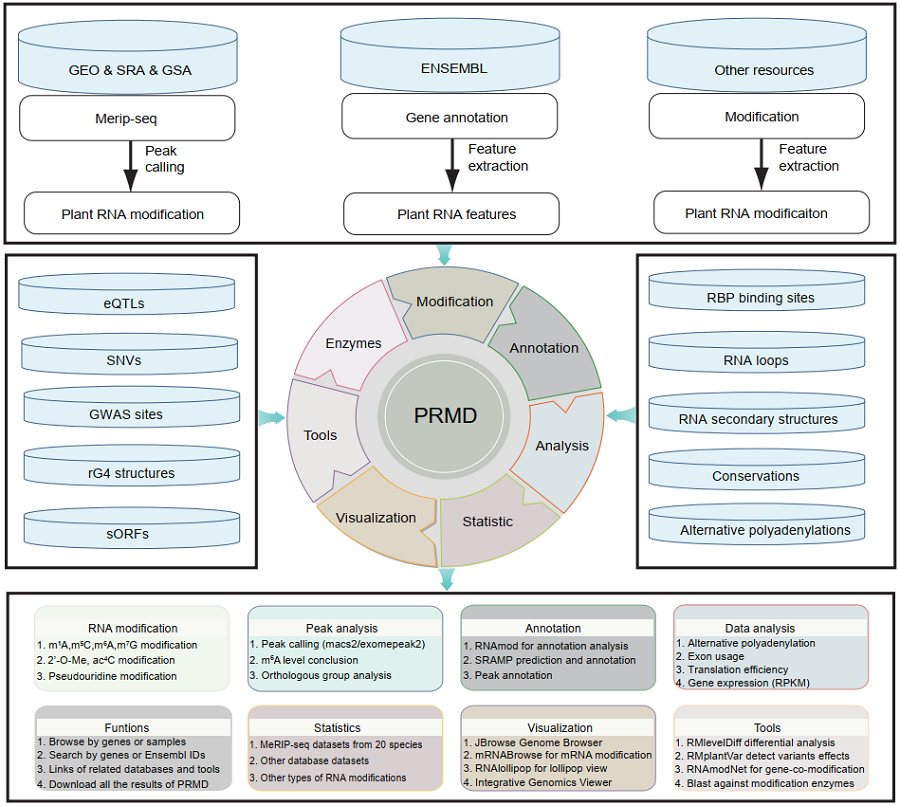
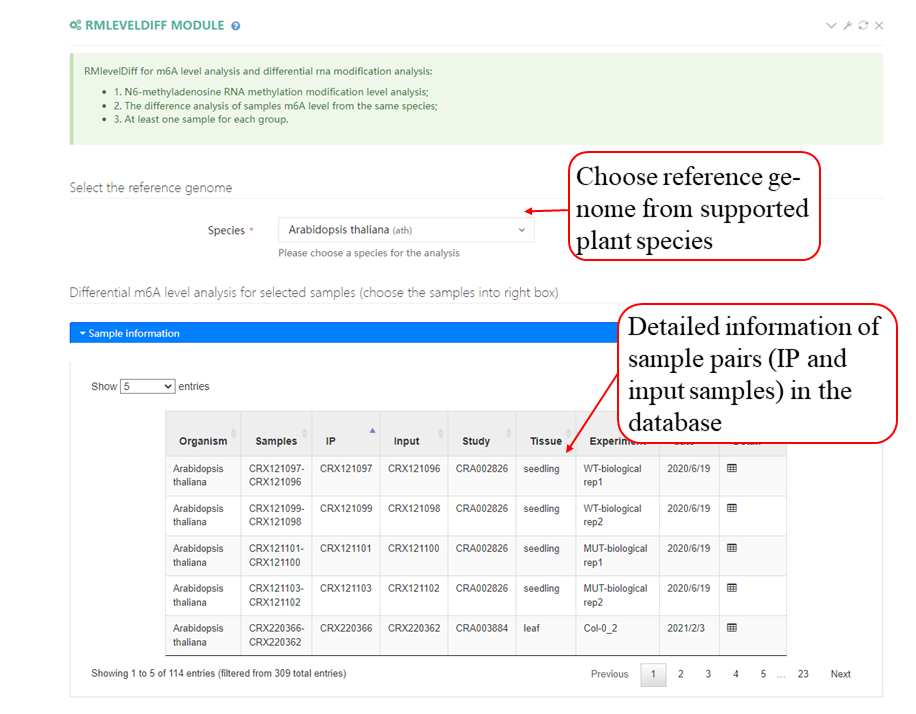
The scheme of PRMD workflow: PRMD provides comprehensive information on RNA modifications. For m6A, 693 MeRIP-seq samples were carefully collected from the SRA and GSA databases for the following 19 plant species: Ae. tauschii (6), A. thaliana (259), B. rapa (24), F. vesca (18), G. max (24), G. arboreum (6), G. hirsutum (18), M. domestica (32), N. benthamiana (18), O. sativa (88), P. fortunei (8), P. patens (6), P. trichocarpa (26), P. vulgaris (6), S. bicolor (22), S. lycopersicum (38), T. aestivum (22), T. dicoccoides (6) and Z. mays (66). In addition, 12 MeRIP-seq samples were generated in this study for O. rufipogon (DXW81), O. sativa ssp. indica (WSSM) and O. sativa ssp. japonica (ZH11). The overall statistical analysis of all datasets indicated the m6A methylation ratios were negatively correlated with the genome size and the number of genes in these plant species. Moreover, PRMD integrated datasets of other types of RNA modifications, such as m1A, m5C, m7G, ac4C, 2′-O-Me and pseudouridine, as well as additional related datasets, including those for eQTLs, SNVs, GWAS sites, rG4 structures, sORFs, RBP binding sites, RNA loops, RNA secondary structures, conservations and APA site information. All datasets were processed through our uniform pipelines. The information was deposited in a MySQL database and displayed in convenient web modules in PRMD. Furthermore, we designed PRMD to enable users to easily visualize and analyze the data in the database.
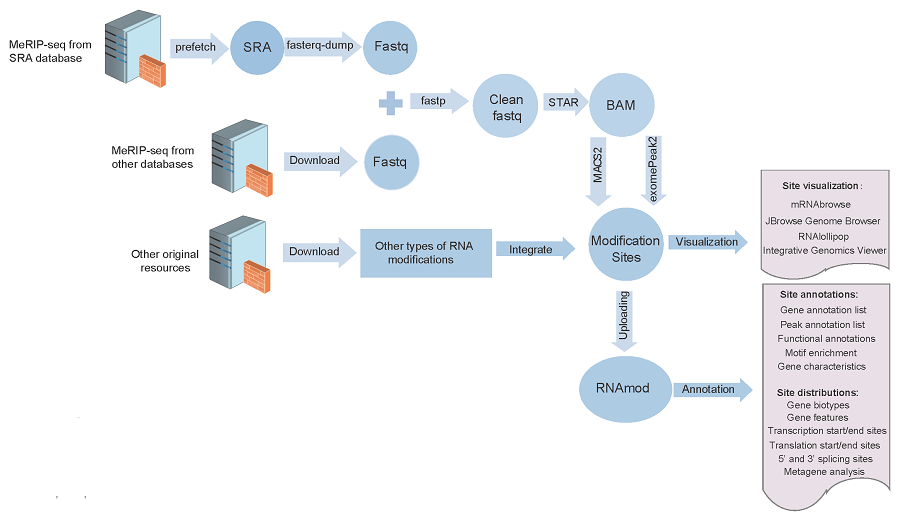
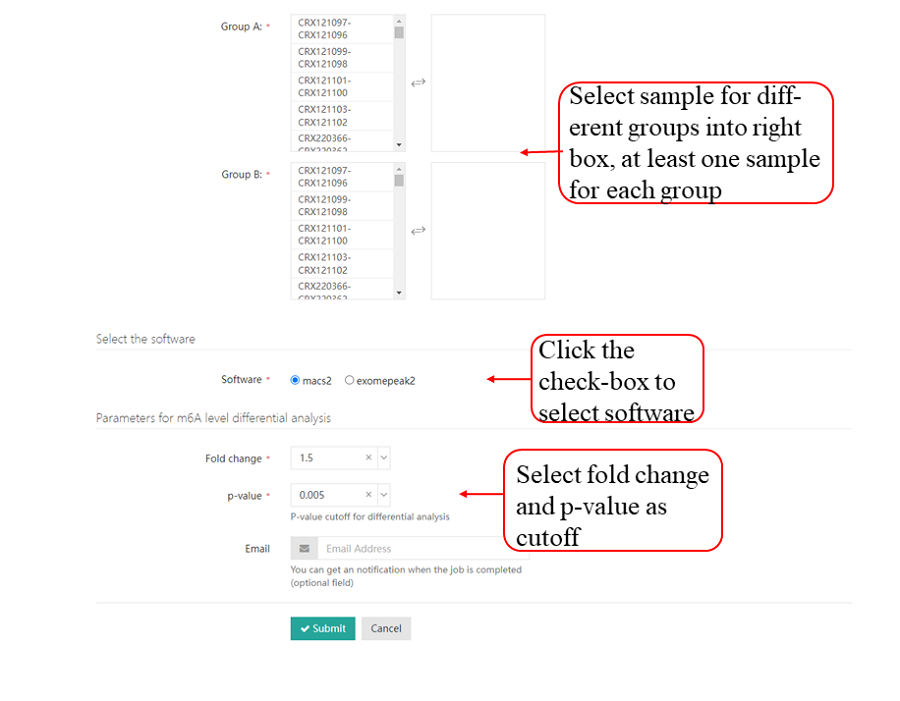
The MeRIP-seq datasets in the SRA format were converted to the FASTQ format using sratoolkit and then the default parameters of fastp were used to trim the adapter sequences and low-quality bases. After filtering the data for quality, STAR was used to map clean reads to the corresponding reference genomes to generate BAM format results. And then using the BAM format genome mapping files of the IP and the input samples, we applied two peak calling strategies. The following parameters of MACS2 were used to identify methylation peaks: --nomodel --extsize 150 -B -n -q 0.05. The R package exomePeak2 (https://github.com/ZW-xjtlu/exomePeak2) was used for peak calling, which involved the same BAM files and GTF files. The parameters were set as follows: fragment_length = 100 binding_length = 25 step_length = 25 pc_count_cutoff = 5 bg_count_cutoff = 50 p_cutoff = 1e-05 peak_calling_mode = exon.
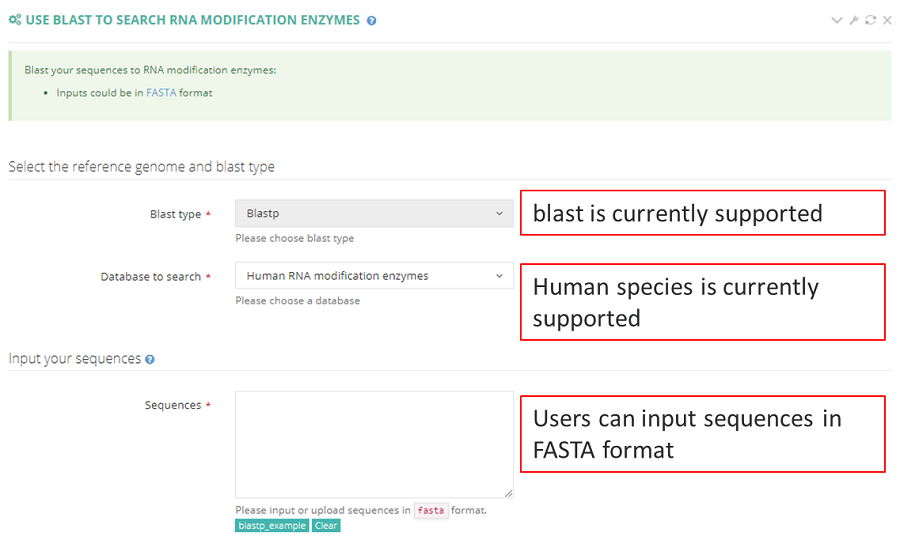
RNAmod (https://bioinformatics.sc.cn/RNAmod), which is an interactive and freely available platform for the annotation and visualization of RNA modifications, was used to annotate the RNA modifications in PRMD. First, RNAmod extracted gene features, such as promoter regions, 5′ and 3′ untranslated regions (UTRs), start codon regions, coding sequence (CDS) regions and stop codon regions, from different annotated reference genomes and then examined gene characteristics, including the GC content, length and minimum free energy. Second, RNAmod mapped all modification sites to different RNA features and calculated coverage values and analyzed metagenes and other annotations. The modified genes were functionally characterized on the basis of Gene Ontology (GO) and KEGG pathways using the clusterProfiler package and according to Reactome pathways using ReactomePA packages.
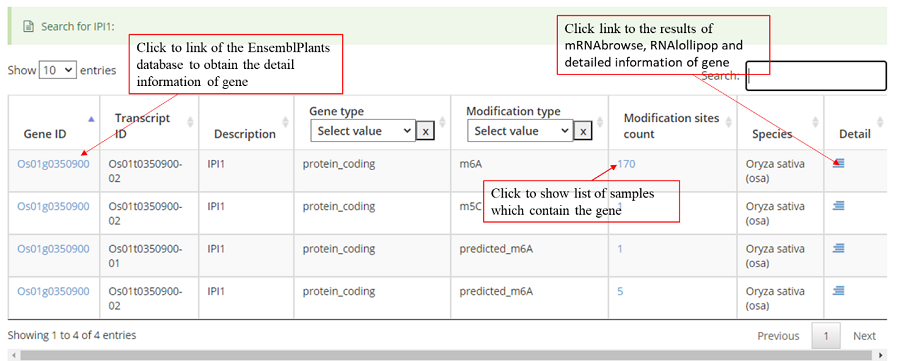
The "Search" module in PRMD allows users to quickly obtain comprehensive information by submitting gene IDs, transcript IDs, gene symbols, sample IDs, study IDs, or PubMed IDs. We also added a quick-search function that supports one query on the PRMD homepage.
The results of the “Search” module revealed that the two transcripts derived from this gene differ in terms of two RNA modifications (m6A and m5C) in PRMD.
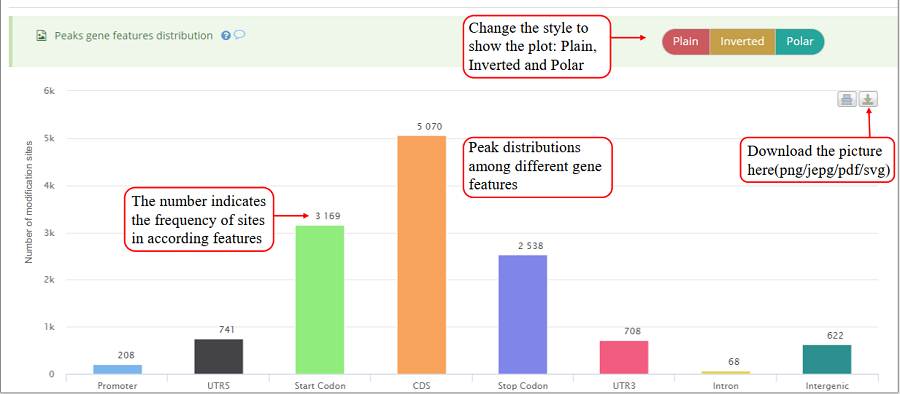
Modification sites distribution on different gene features, including Promoter, 5'UTR (UTR5), CDS, 3'UTR (UTR3), Start codon, Stop codon, Intron and Intergenic region. Y-axis denotes the frequency of sites (number of sites) while x-axis represents different gene features. The numbers on the bar indicates the frequency of sites distributed on according gene feature. The plot can be showed in three different forms: 'Plain', 'Inverted' and 'Poplar' and the plot can be exported in png, jpeg, pdf and svg formats. It is noted that the stop codon is overlapped with 3'UTR and CDS.
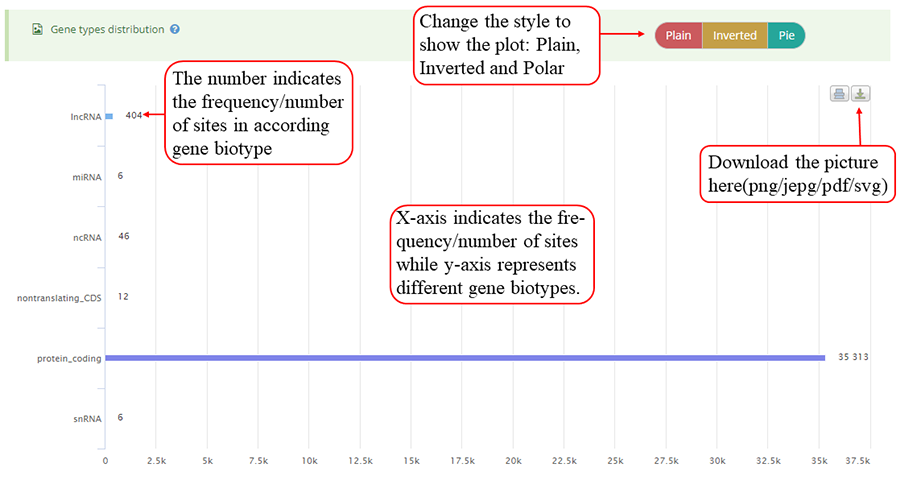
Modification sites distribution on different gene biotypes, such as protein coding gene, lncRNA, pseudogenes, rRNA, and miRNA. Y-axis indicates the frequency of sites (number of sites) while x-axis represents different gene biotypes. The numbers on the bar indicates the frequency of sites distributed on according gene biotype. The plot can be showed in three different forms: 'Plain', 'Inverted' and 'Pie' and can be downloaded in png, jepg, svg and PDF formats.
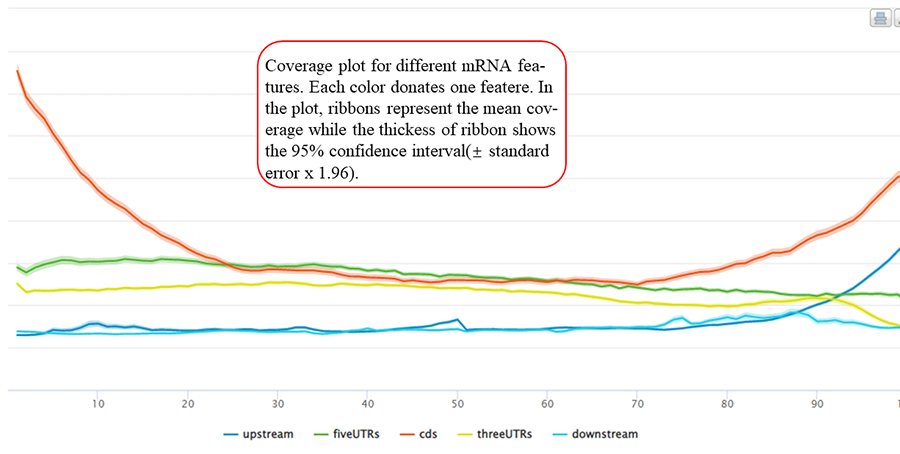
Coverage plot for modification sites overlapping with different mRNA features. After excluding the features shorter than specific length, each gene feature is divided into bins (100 bins by default) in equal size. The value of the length threshold equals to the number of bins. The number of sites distributed in each bin is counted and the mean coverage is then calculated among all the according features. X-axis represents the bins (5' → 3' direction) while Y-axis indicates the mean coverage. In the plot, ribbons represent the mean coverage while the thickness of ribbon shows the 95% confidence interval (±standard error x 1.96).
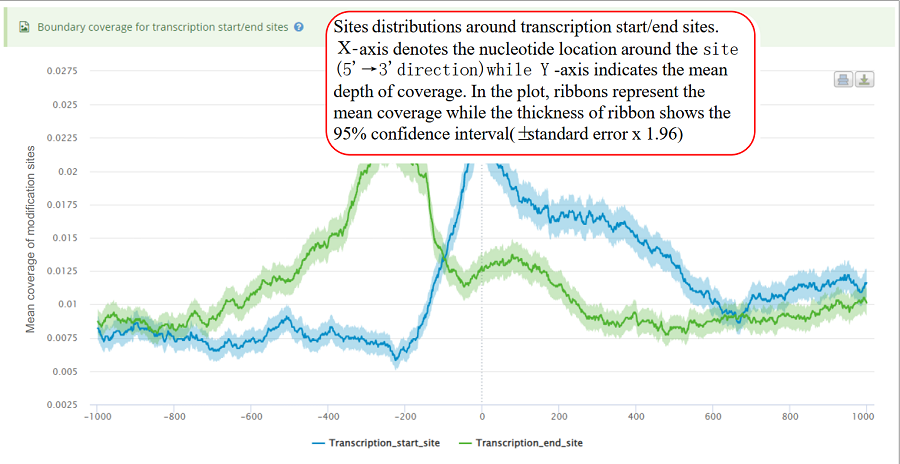
Coverage plot for modification sites around transcription start sites and translation end sites. The number of sites for each location is counted around the flank regions (upstream and downstream,1000 bp flank size in default). The mean depth of coverage for that location is then calculated for all the genes. X-axis represents the nucleotide location around the site (5' → 3' direction) while Y-axis is the mean depth of coverage. In the plot, ribbons represent the mean coverage while the thickness of ribbon shows the 95% confidence interval (±standard error x 1.96).
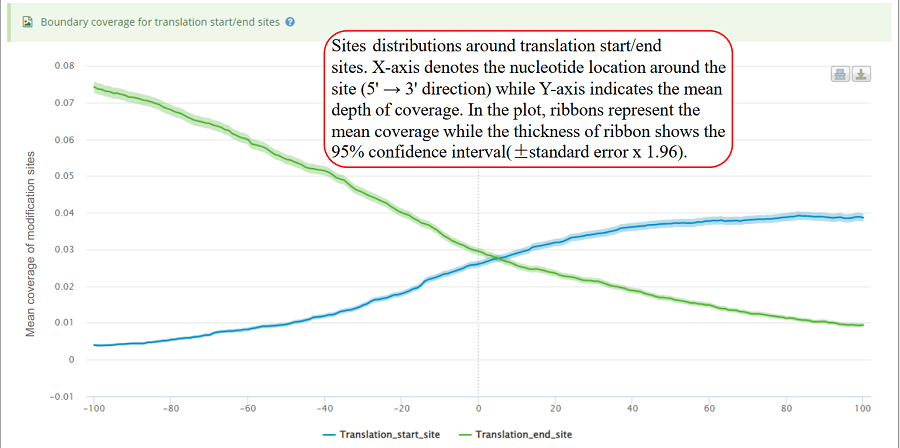
Coverage plot for modification sites around translation start sites (TSS) and translation end sites (TES). The number of sites for each nucleotide location is counted around the flank regions (upstream and downstream) of TSS/TES on the transcript. The mean depth of coverage is then calculated for all the genes. X-axis represents the nucleotide location around the site (5' → 3' direction) while Y-axis is the mean depth of coverage. In the plot, ribbons represent the mean coverage while the thickness of ribbon shows the 95% confidence interval (±standard error x 1.96).
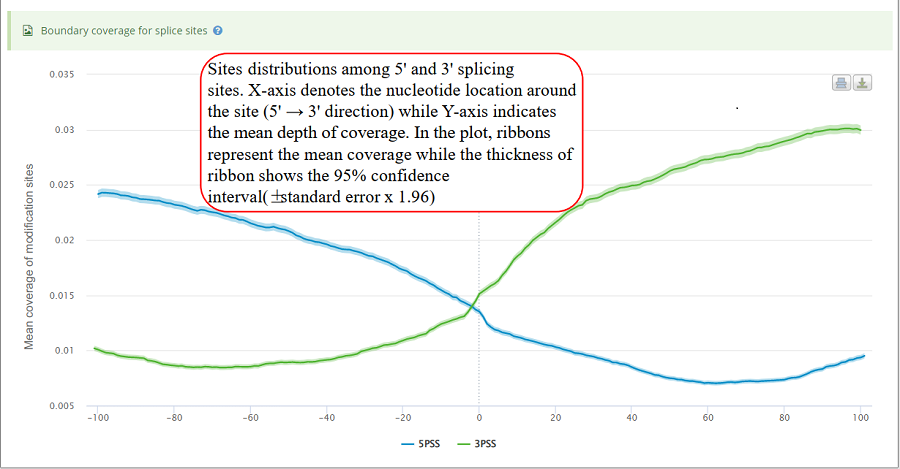
Coverage plot for modification sites around 5' splice sites and 3' splice sites. For the coverage analysis around splice sites, the number of sites in each location is counted. Then, mean depth coverage for specific location is calculated for all the genes. X-axis represents the nucleotide location around the splice site (5' → 3' direction) while Y-axis indicates the mean depth of coverage. In the plot, ribbons represent the mean coverage while the thickness of ribbon shows the 95% confidence interval (±standard error x 1.96).
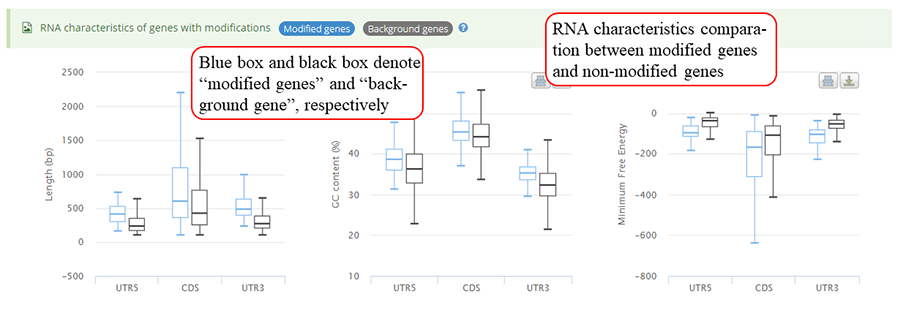
mRNA characteristics statistics between genes with modifications and other background genes. Y-axis in three plots (from left to right) represents length, GC content and minimum free energy (MFE), respectively. The blue bar and black bar represent modified gene and other background genes, respectively.
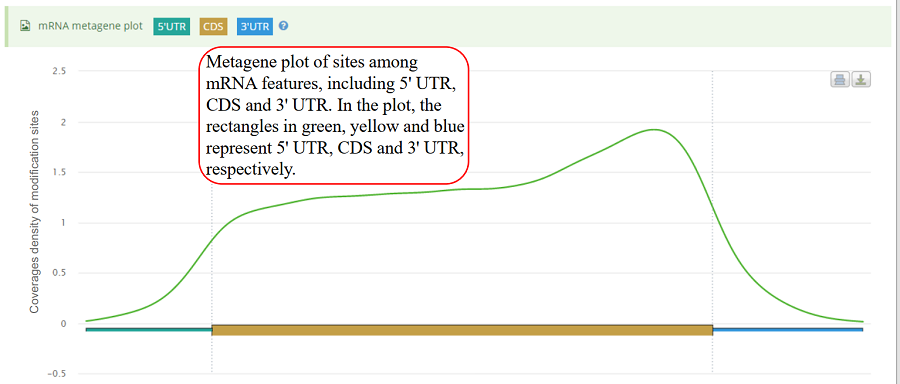
mRNA metagene plot. After excluding the gene with any of the mRNA features (5'UTR, CDS and 3'UTR) shorter than specific length, each mRNA feature was divided into bins with equal size (100 bins by default). The value of the length threshold equals to the number of bins. The number of sites distributed in each bin is counted. Then,the mean coverage in each bin is calculated among all the genes. The rectangles in green, yellow and blue represent 5'UTR, CDS and 3'UTR, respectively. Y-axis denotes the density distribution of coverages.
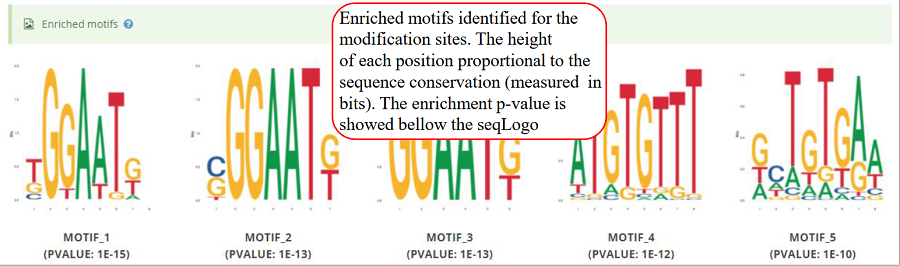
Enriched motifs detect by Homer for the modification sites. The seqLogo plots for enriched motifs (top 5 as default) are showed, in which, the logo use a stack of letters to represents columns of the alignment and the height of each stack proportional to the sequence conservation (measured in bits) at that position. The enrichment p-value is showed bellow according seqLogo plot.
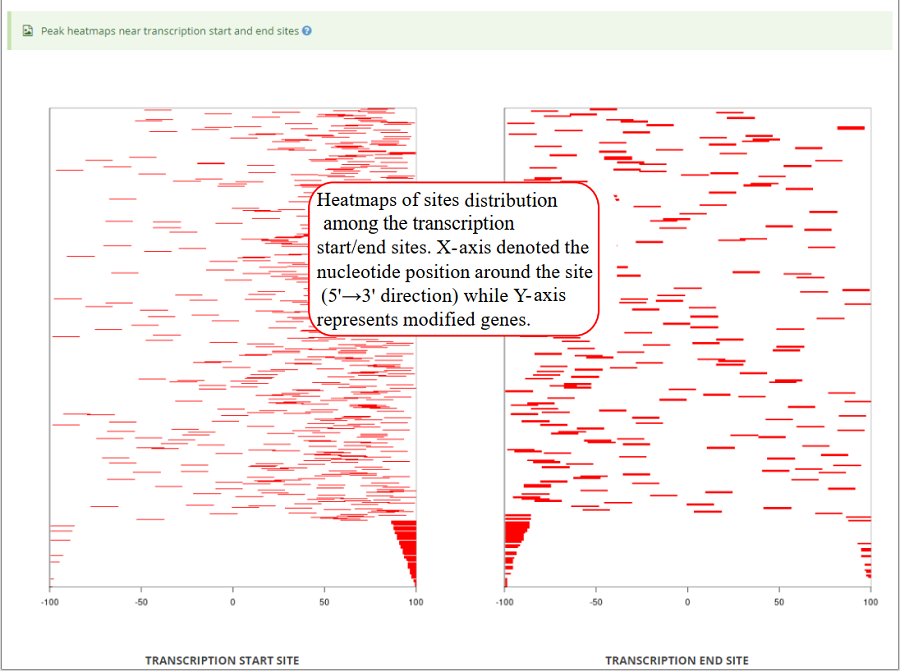
Heatmap of modification sites around transcription start sites and transcription end sites (genomic regions). X-axis represents the nucleotide position around the site (5' → 3' direction) while Y-axis represents all modified genes which is sorted by the number of modifications in each gene. In the plot, the red color scale evaluates the depth of coverage.
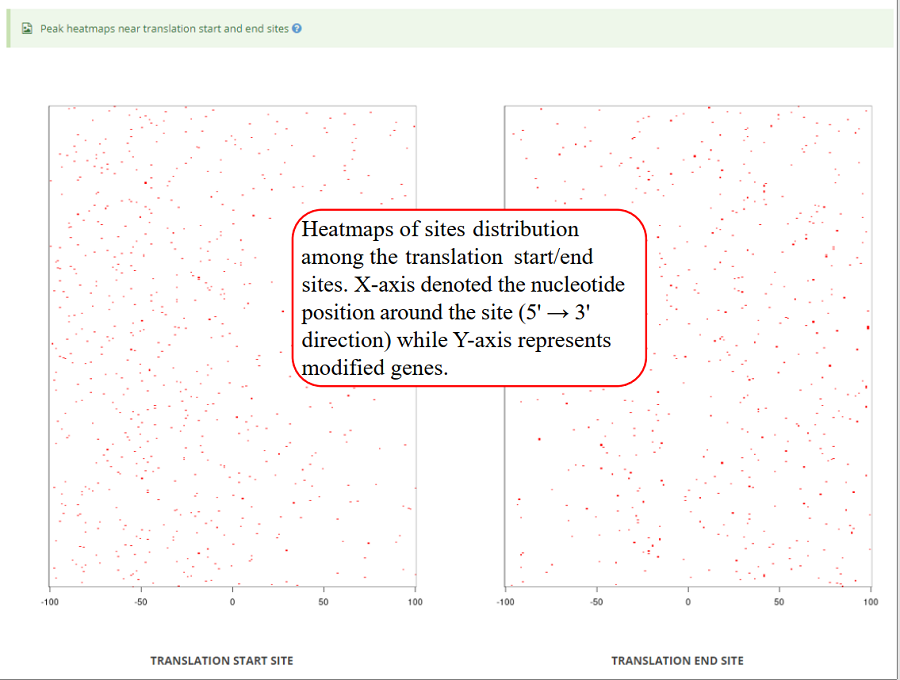
Heatmap of modification sites among translation start sites and translation end sites (transcriptic regions). X-axis represents the nucleotide position around the site (5' → 3' direction) while Y-axis represents all modified genes which is sorted by the number of modifications in each gene. In the plot, the red color scale evaluates the depth coverages.
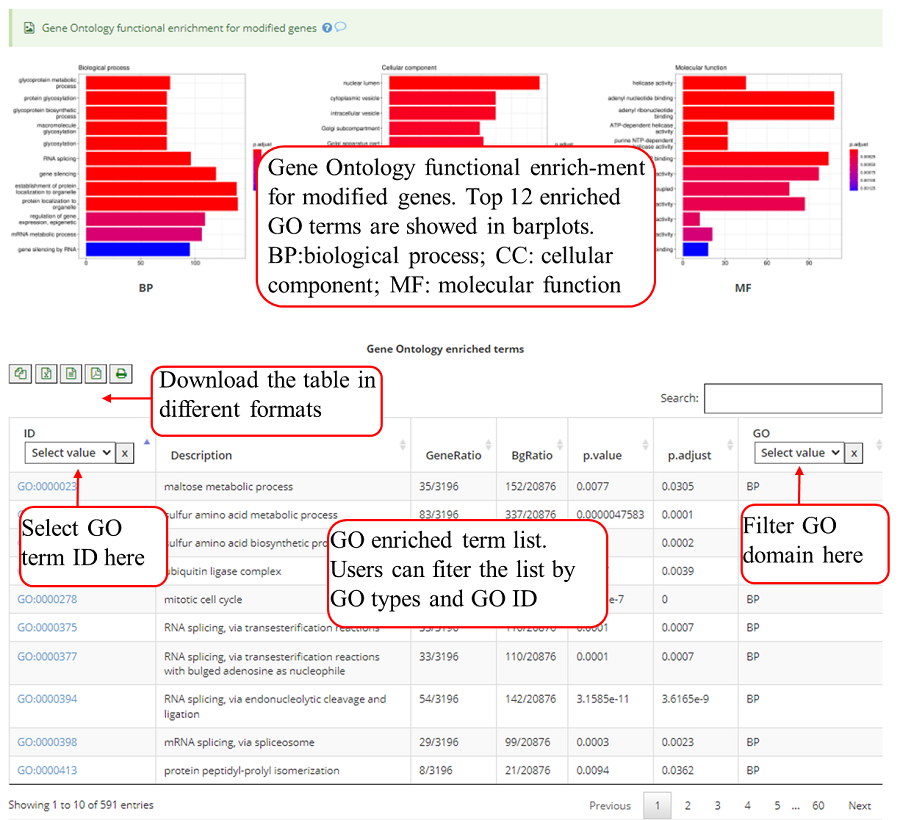
Gene Ontology (GO) functional enrichment for genes with modifications. Top 12 enriched terms are show in the bar plots. Y-axis in the bar plot indicates number of genes in according GO term. The color scale represent the enrichment p-value. All the enriched terms in three GO domains (biological process, cellular component and molecular function) are contained in one table and use filter in 'GO' column to select specific GO domain terms. A blank picture indicates that there are no significant enriched terms for that GO domain.
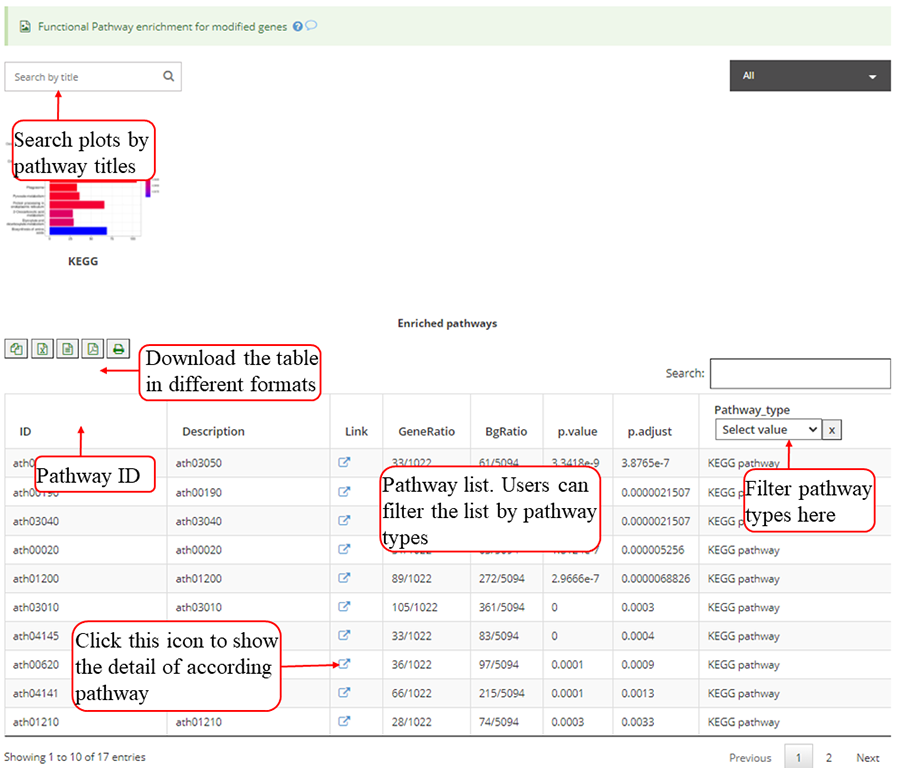
Functional pathway enrichments for genes with modifications, which include KEGG for all 20 species, Top 12 enriched pathways are show in the barplots. Y-axis in the barplot indicates number of genes in according pathway category. The color scale represent the enrichment p-value. All the enriched pathway in different pathways are contained in one table and use filter in 'Type' column to select specific pathway. If there is no data for specific pathway category, there will be no picture for that pathway. Beside, a blank picture indicates that there is no significant enriched pathway for that pathway category.
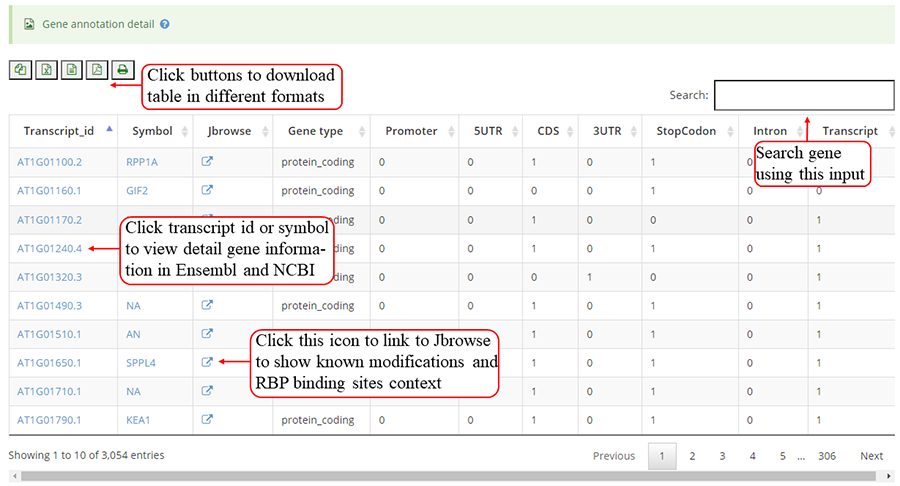
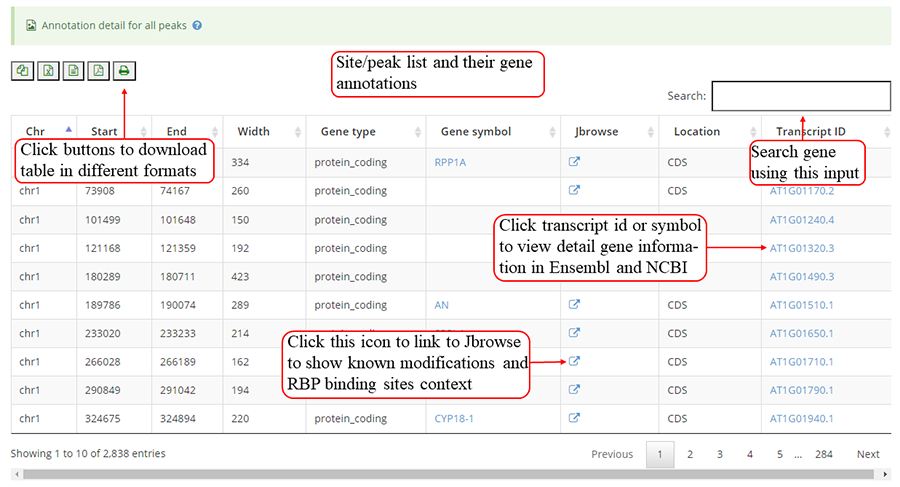
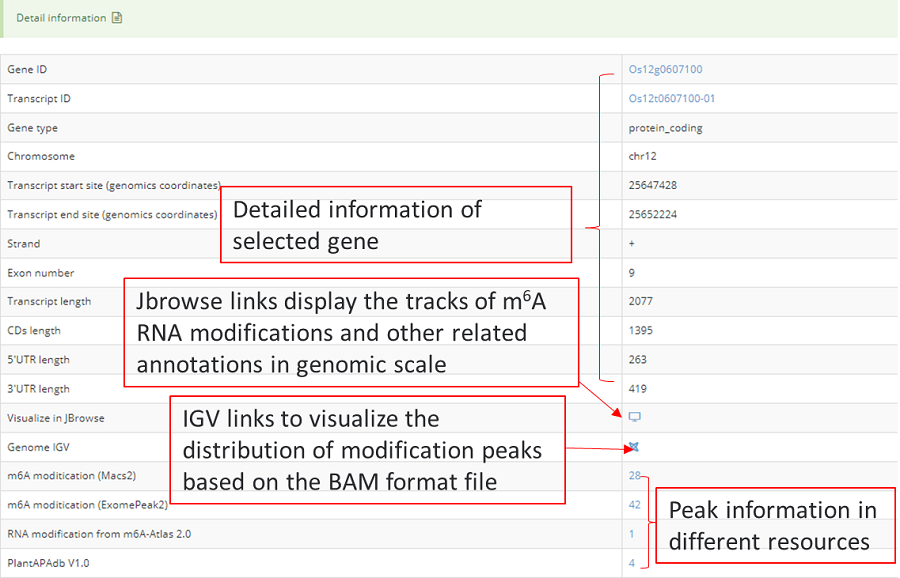
The detail list of genes containing modification sites. The gene information include transcript ID, gene symbol and gene biotype (gene_type). It is note that 'Stop codon' region is overlapped with 3'UTR and CDS. Click icon to compare the modifications in these genes with known modifications and RBP binding sites in JBrowse and click the transcript_ID to show the detailed gene information in Ensembl database.
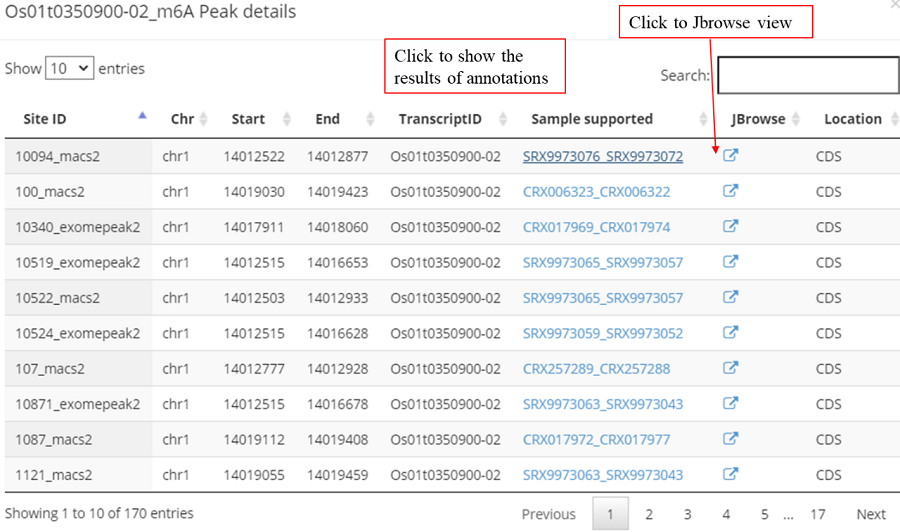
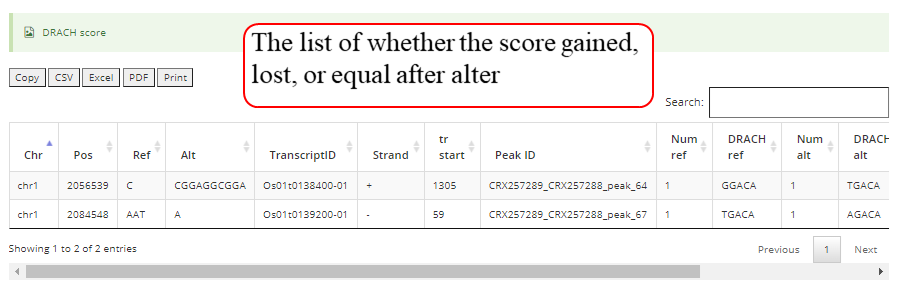
Detail annotation list of sites. The gene information for the peak include transcript ID, gene symbol, gene biotype (gene type) and location on according gene. Click icon to compare the modification with other known modifications and RBP binding sites in JBrowse while click the transcript ID to show the detailed gene information in Ensembl database.
The m6A peak sites were identified by our own uniform pipeline based on the MeRIP-seq datasets, while all the potential m6A sites were predicted by the m6A prediction software. SRAMP requires nucleotide sequence only for running a prediction. There are two prediction modes available, each accepts different kinds of sequences, which are specified below:
Full transcript mode: This mode is recommended when running predictions on both coding and non-coding RNAs, due to its superior accuracy. Note that in this mode, genomic sequence of the full transcript (with introns) rather than mature mRNA/cDNA sequences should be used.
Mature mRNA mode:This prediction mode is an alternative solution for users who do not have genomic sequences at hand, and its performance is still competitive. This model works on mature mRNA (cDNA) sequences and CANNOT predict m6A sites in introns.
The thresholds for very high/high/moderate/low confidence m6A sites correspond to the thresholds achieved 99%/95%/90%/85% specificities (in other words, had 5%/10%/15% false positive rate) on cross-validation tests, respectively. These predicted m6A sites were as a supplementary data set in the PRMD database.
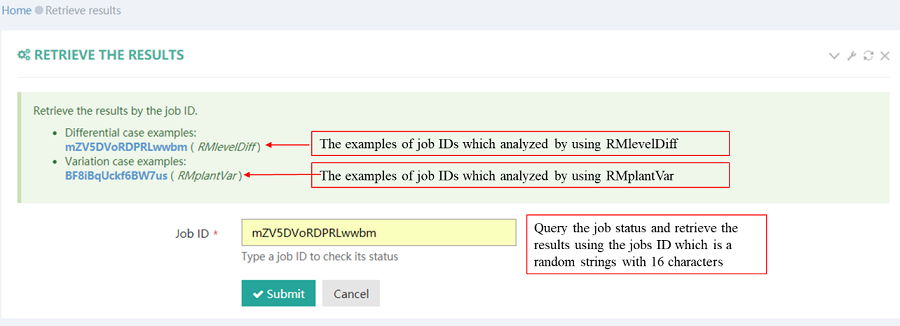
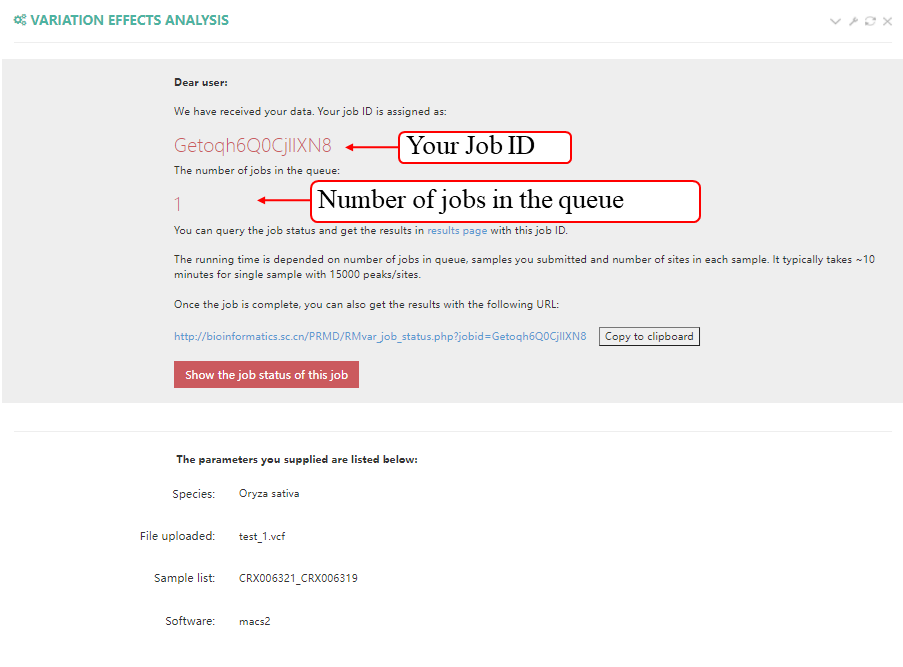
After job is submitted, the web server will give the user a Job ID, and display the running progressing.
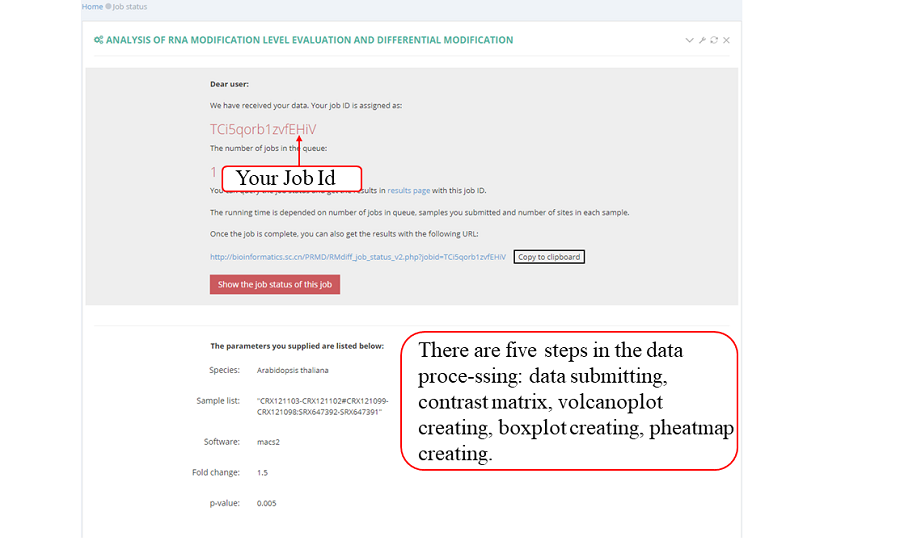
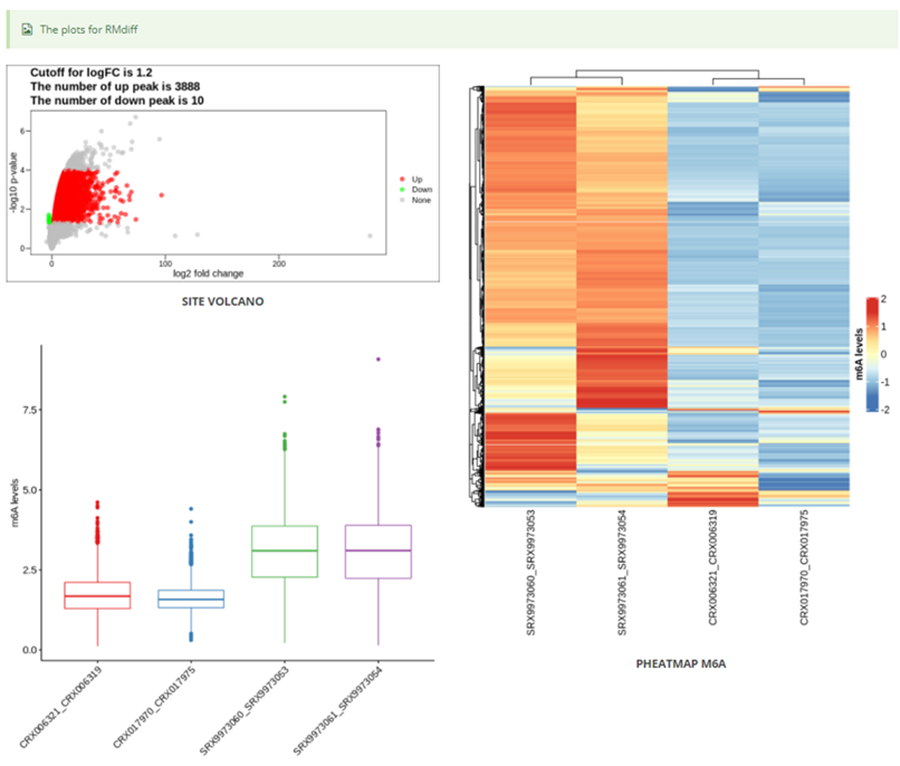
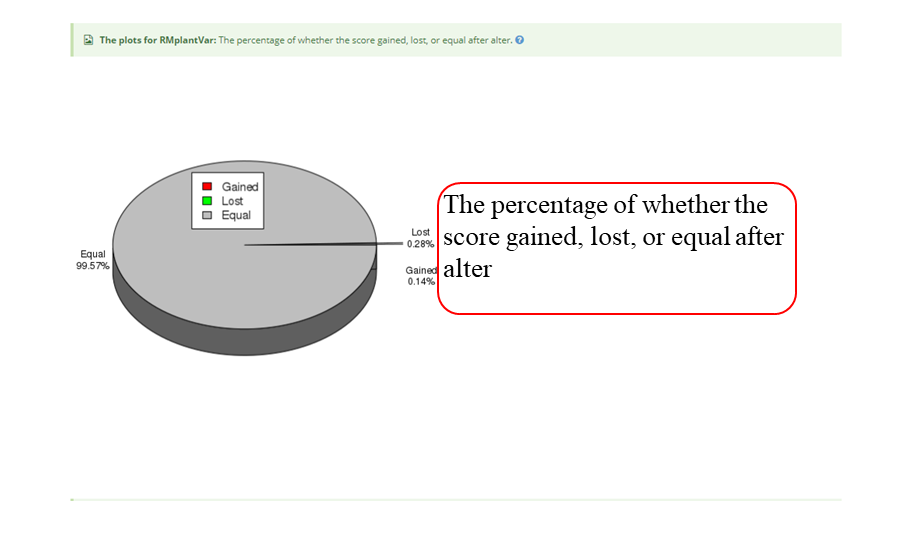
The page while redirect to the result page when the job completed. In volcano plot, each point represents a site, red represents up-regulated peak and green represents down-regulated peak according to cutoff giving by users. X-axis represents log2 fold change, Y-axis represents –log10(p-value). In box plot, X-axis represents samples, Y-axis represents log2(m6A levels + 1). In heatmap, X-axis represents the samples, Y-axis represents all modified sites which is clusterd by the m6A modification levels.
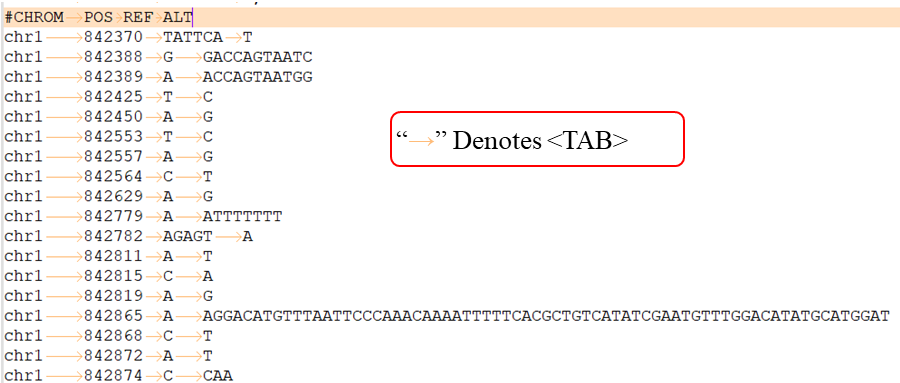
VCF lines have four required fields:
CHROM - chromosome: An identifier from the reference genome or an angle-bracketed ID String ("< ID >") pointing to a contig in the assembly file (cf. the ##assembly line in the header). All entries for a specific CHROM should form a contiguous block within the VCF file. (String, no whitespace permitted, Required).
POS - position: The reference position, with the 1st base having position 1. Positions are sorted numerically, in increasing order, within each reference sequence CHROM. It is permitted to have multiple records with the same POS. Telomeres are indicated by using positions 0 or N+1, where N is the length of the corresponding chromosome or contig. (Integer, Required).
REF - reference base(s): Each base must be one of A,C,G,T,N (case insensitive). Multiple bases are permitted. The value in the POS field refers to the position of the first base in the String. For simple insertions and deletions in which either the REF or one of the ALT alleles would otherwise be null/empty, the REF and ALT Strings must include the base before the event (which must be reflected in the POS field), unless the event occurs at position 1 on the contig in which case it must include the base after the event; this padding base is not required (although it is permitted) for e.g. complex substitutions or other events where all alleles have at least one base represented in their Strings. If any of the ALT alleles is a symbolic allele (an angle-bracketed ID String "< ID >") then the padding base is required and POS denotes the coordinate of the base preceding the polymorphism. Tools processing VCF files are not required to preserve case in the allele Strings. (String, Required).
ALT - alternate base(s): Comma separated list of alternate non-reference alleles. These alleles do not have to be called in any of the samples. Options are base Strings made up of the bases A,C,G,T,N,*, (case insensitive) or an angle-bracketed ID String ("< ID >") or a breakend replacement string as described in the section on breakends. The ‘*’ allele is reserved to indicate that the allele is missing due to a upstream deletion. If there are no alternative alleles, then the missing value should be used. Tools processing VCF files are not required to preserve case in the allele String, except for IDs, which are case sensitive. (String; no whitespace, commas, or angle-brackets are permitted in the ID String itself).
After job is submitted, the web server will give the user a Job ID, which can be used to retrieve the results.
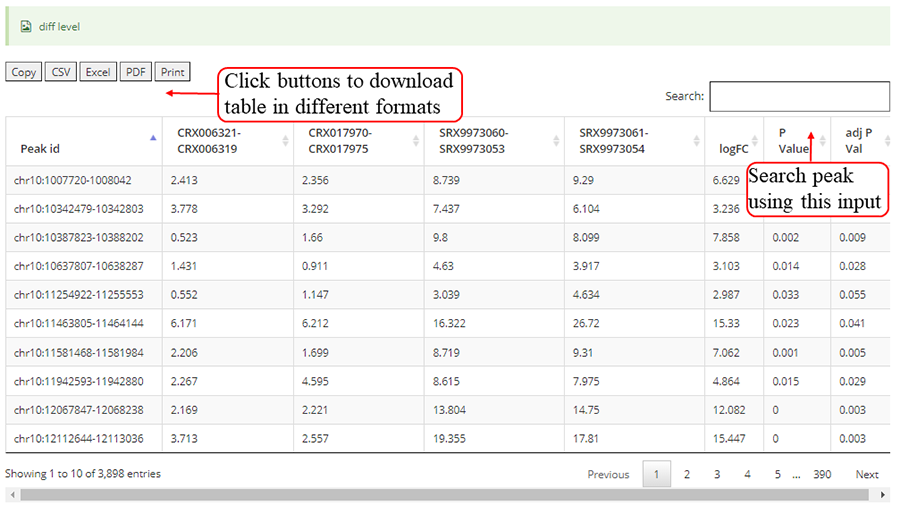
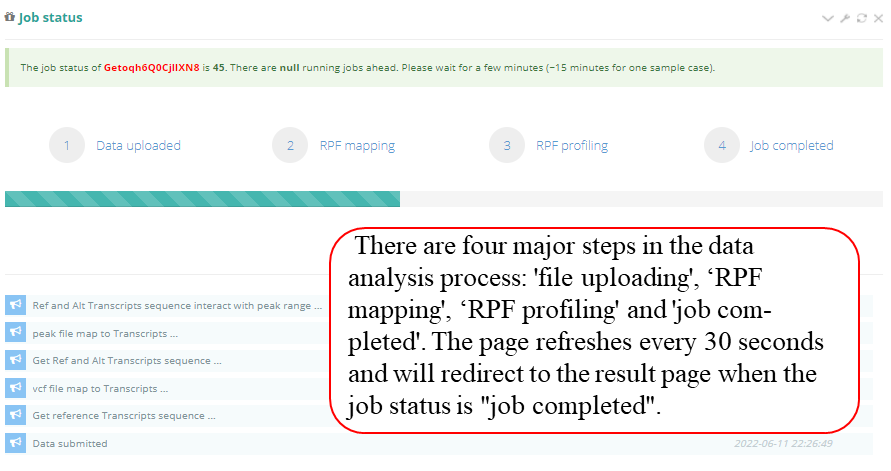
Query the job status. There are four major steps in the data analysis process: RPF mapping', ‘RPF profiling' and 'job completed'. The page refreshes every 30 seconds and will redirect to the result page when the job status is "job completed". In result pages, all the results are showed in high-quality interactive figures and tables. The figures generated by highcharts.js can be downloaded in PNG, JPG, PDF and SVG formats while the table formated by DataTables.js can be download in CSV, Excel and PDF formats.
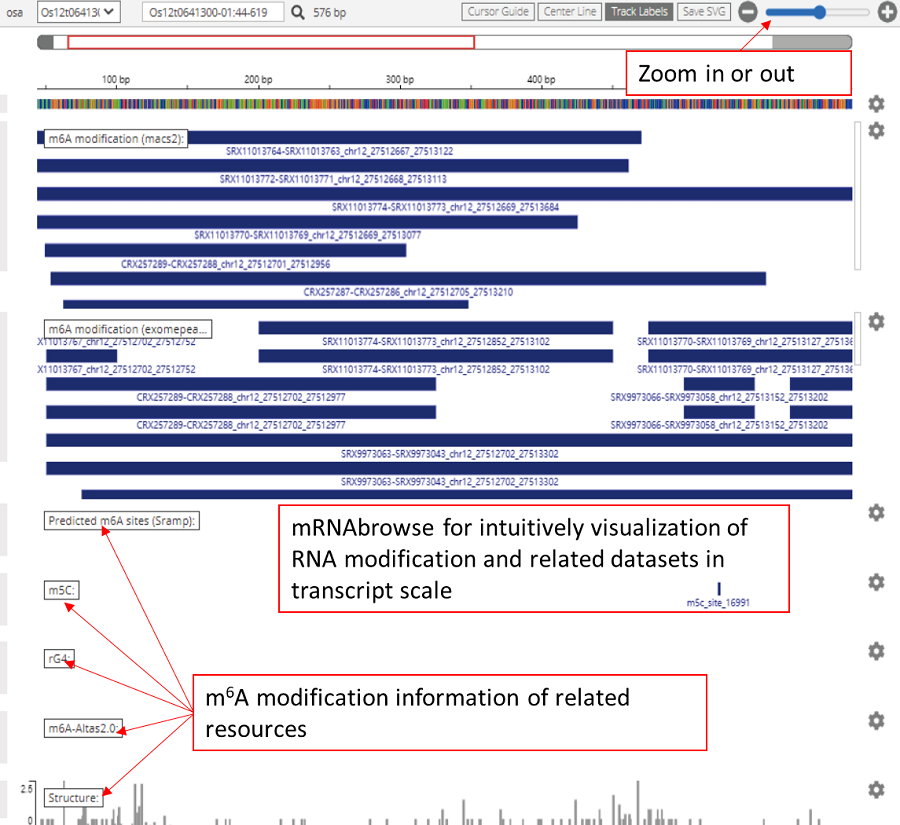
mRNAbrowse was designed for the intuitive visualization of RNA modifications and related datasets (transcript scale), including modification sites determined by nanopore sequencing and miCLIP-seq, sequence conservation, GWAS sites, miRNA target sites, APA sites, RBP binding sites, RNA secondary structures, rG4 structures, R-loop elements, sORFs and other types of modifications. In mRNAbrowse, users can zoom in and out using buttons in the upper right corner to visualize the modification site sequence context.
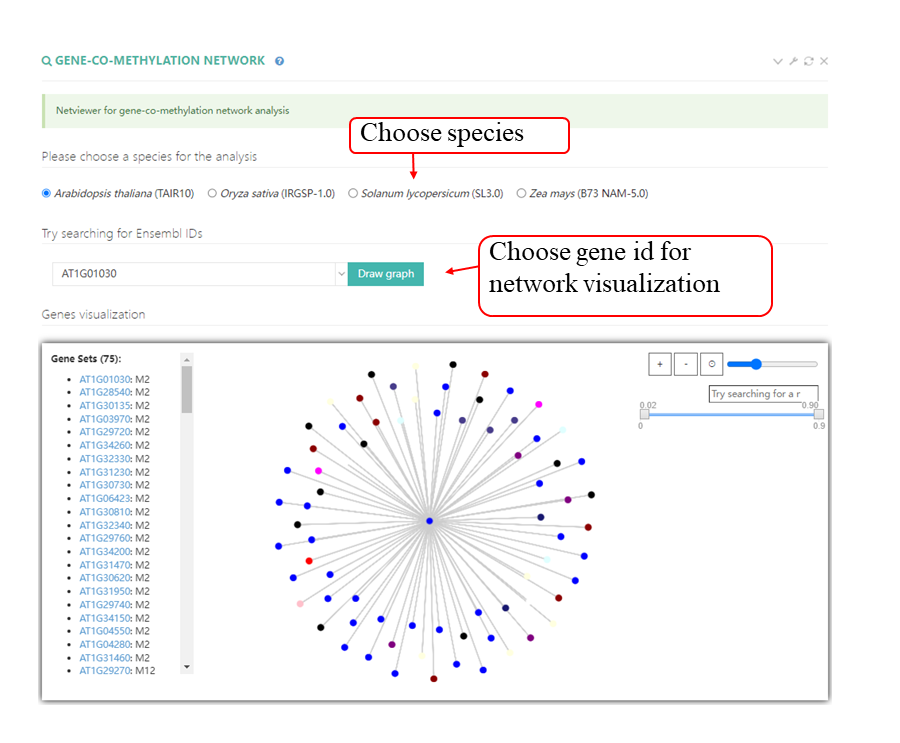
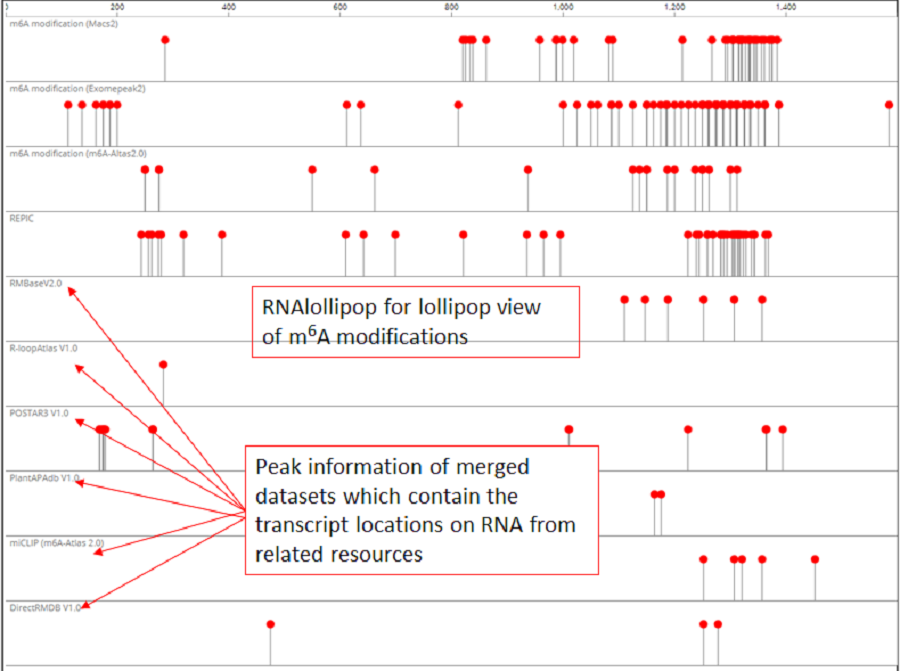
RNAlollipop was designed to establish lollipop views of the modifications and other datasets in PRMD. For each dataset type, all of the datasets were merged to enable users to intuitively compare the RNA modifications with other elements at the same location from different sources. JBrowse was integrated to visualize the modification sites and other information (genome scale).
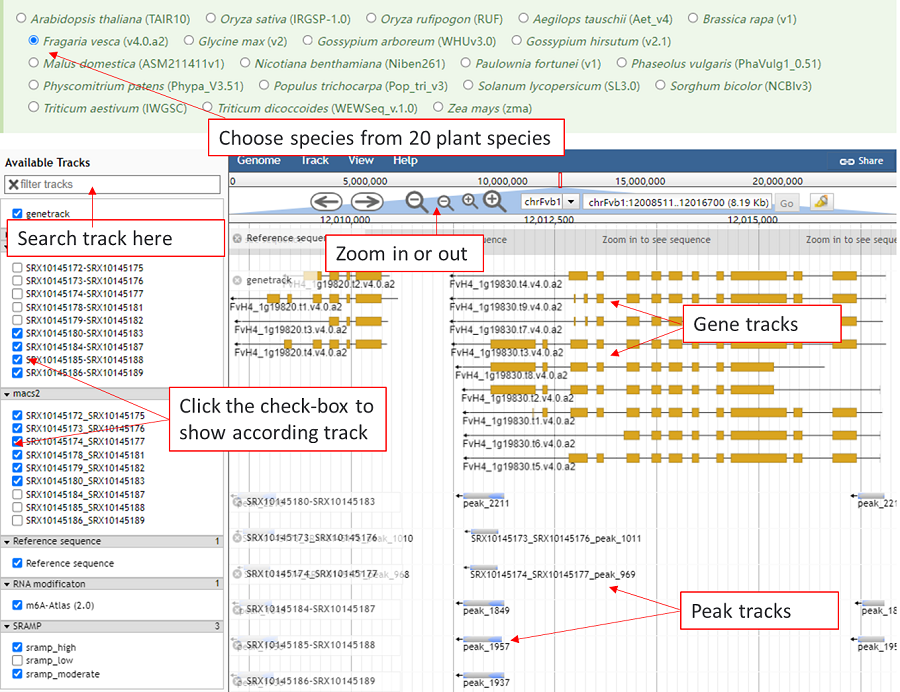
Jbrowse is used to show the known modification sites and related data sites. The tracks are organized into three groups: basic genome information tracks, RNA modification tracks and RBP binding-site tracks. The track of user's modification are showed automatically and click the checkbox in left window to show other tracks. Meanwhile, the tracks can be searched by inputing the modification type or RBP name.
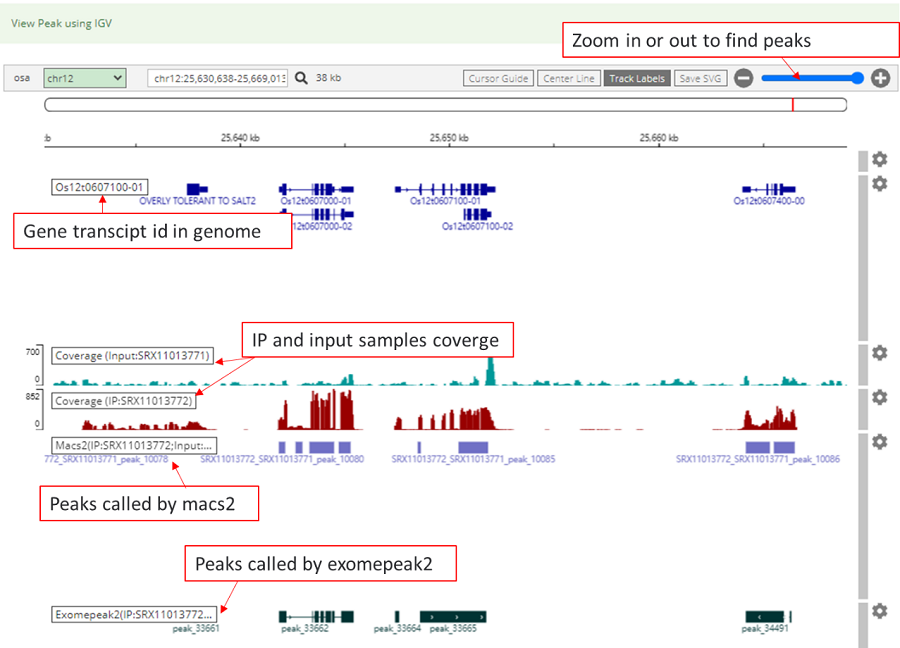
The Integrative Genomics Viewer (IGV), which located in detail information under the view of mRNAbrowse and RNAlollipop, with hyperlinks provided in the table with specific details and information, was integrated to visualize the distribution of the modification peaks on the basis of the BigWig files of the input and IP samples in the genome. Users can intuitively check the read coverage of the modification peaks called by MACS2 and exomePeak2 among different samples.
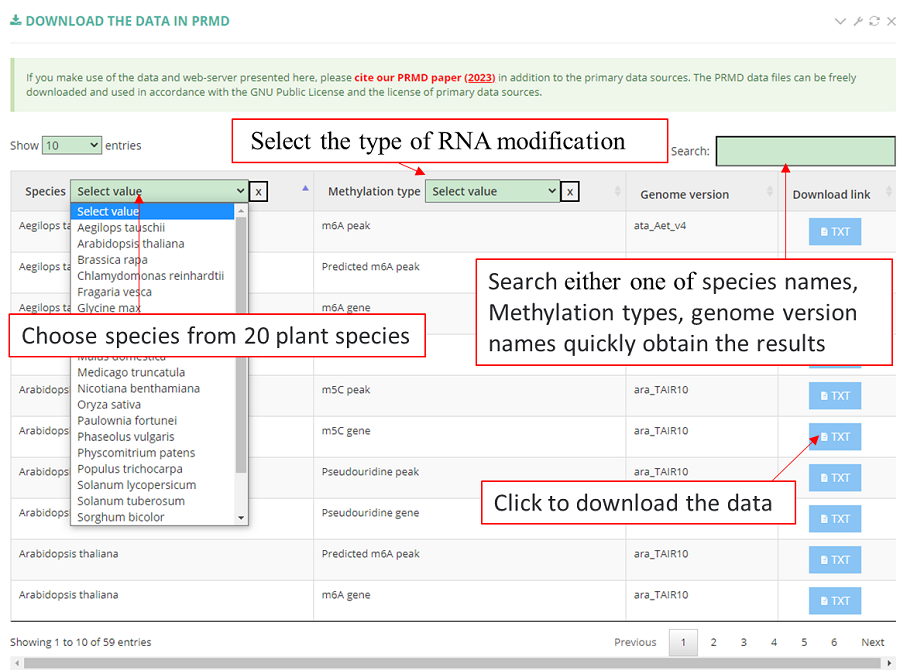
The "Download" module, which provides the datasets of different types of RNA modifications and different species, with users able to download data with one click depending on the species and type of RNA modification.
Web resources have a dependency on internet browsers, and different browsers use different kernels with different approaches. We recommend that users access our database through a Firefox, google or Microsoft Edge browser.
| Browser name | Version |
|---|---|
| Internet Explorer | 11 |
| Chrome | 70.0.3538.80 |
| Firefox | 116.0.1 |
| Microsoft Edge | 112.0.1722.48 |
| safari | 5.1.10 |
| 360 | 14.1.1116.0 |
| Opera | 101.0.4843.43 |

