Welcome to PRMD
The scope and function of RNA modifications in model plant systems have been an intense focus of study with the discovery of an increasing number of novel RNA modifications in recent years. Researchers have gradually realized that RNA modifications, especially N6-methyladenosine (m6A) RNA methylation, one of the most abundant and studied RNA modifications in plants, plays an important role in physiological and pathological processes. These modifications alter the structure of RNA, affecting its molecular complementarity, and binding to specific proteins, resulting in a variety of physiological effects, such as messenger RNA (mRNA) splicing and export, translation and stability. With more and more discoveries and studies of RNA modifications in plants, annotating RNA modifications and their target gene become very essential. However, a convenient and integrated database which focuses on comprehensive annotations, intuitive visualization of plant RNA modifications and other related regulation data is still lack.
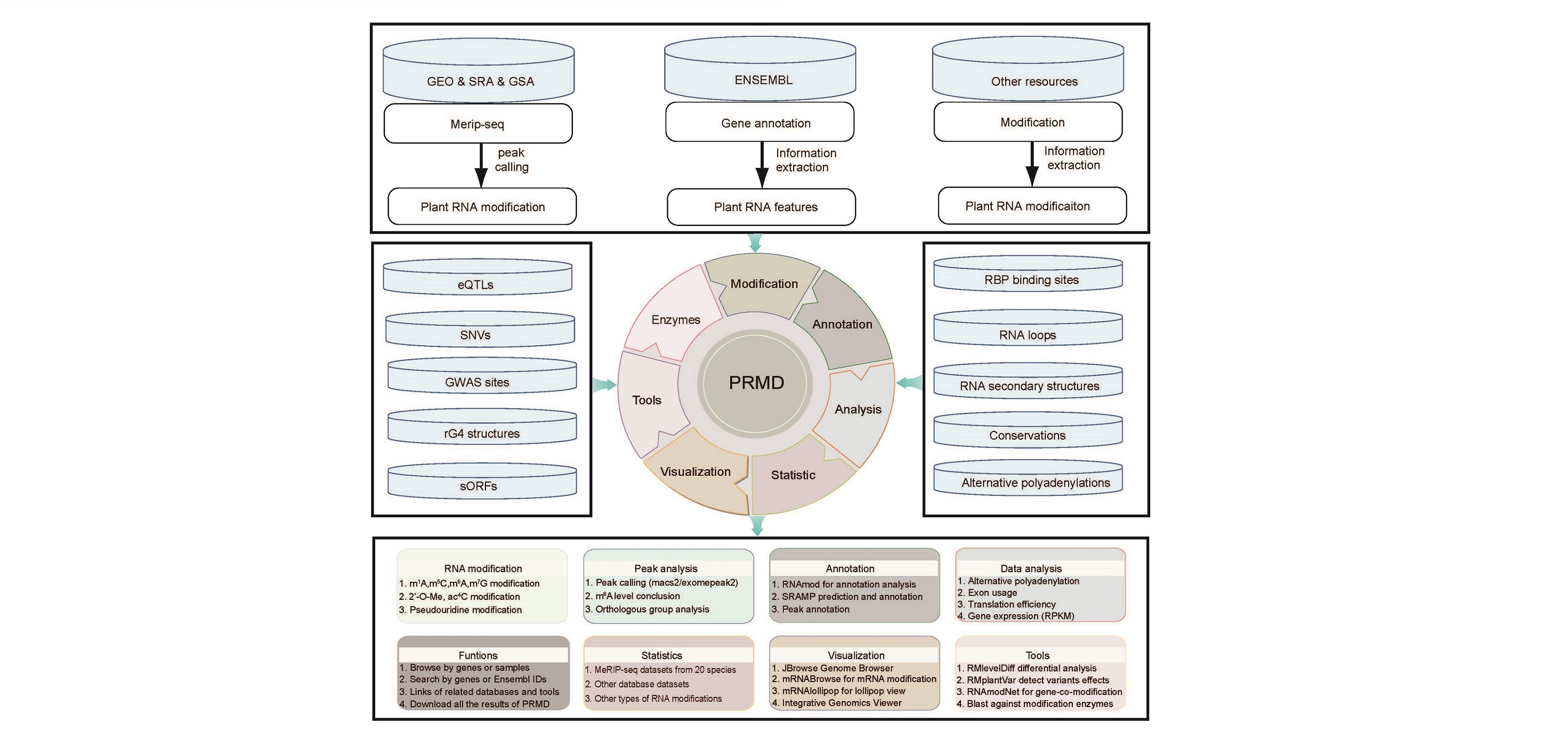
Here, we developed the Plant RNA Modification Database (PRMD), which was established primarily to facilitate RNA modification researches, containing 20 plant species and providing an intuitive interface to display the information. Besides, PRMD provided convenient visualization and functional analysis tools, such as RMlevelDiff, RMplantVar, RNAmodNet and Blast to perform different analyses, and mRNAbrowse, RNAlollipop, JBrowse and Integrative Genomics Viewer (IGV) to visually display the analysis and statistical results. PRMD is completely free and open to users, which provides great convenience for the rapid development and promotion of research on RNA modifications in plants.
The output of PRMD includes:
- The overall statistic of different mRNA modifications features across different species
- The overall statistic of modifications across different RNA features and biotypes
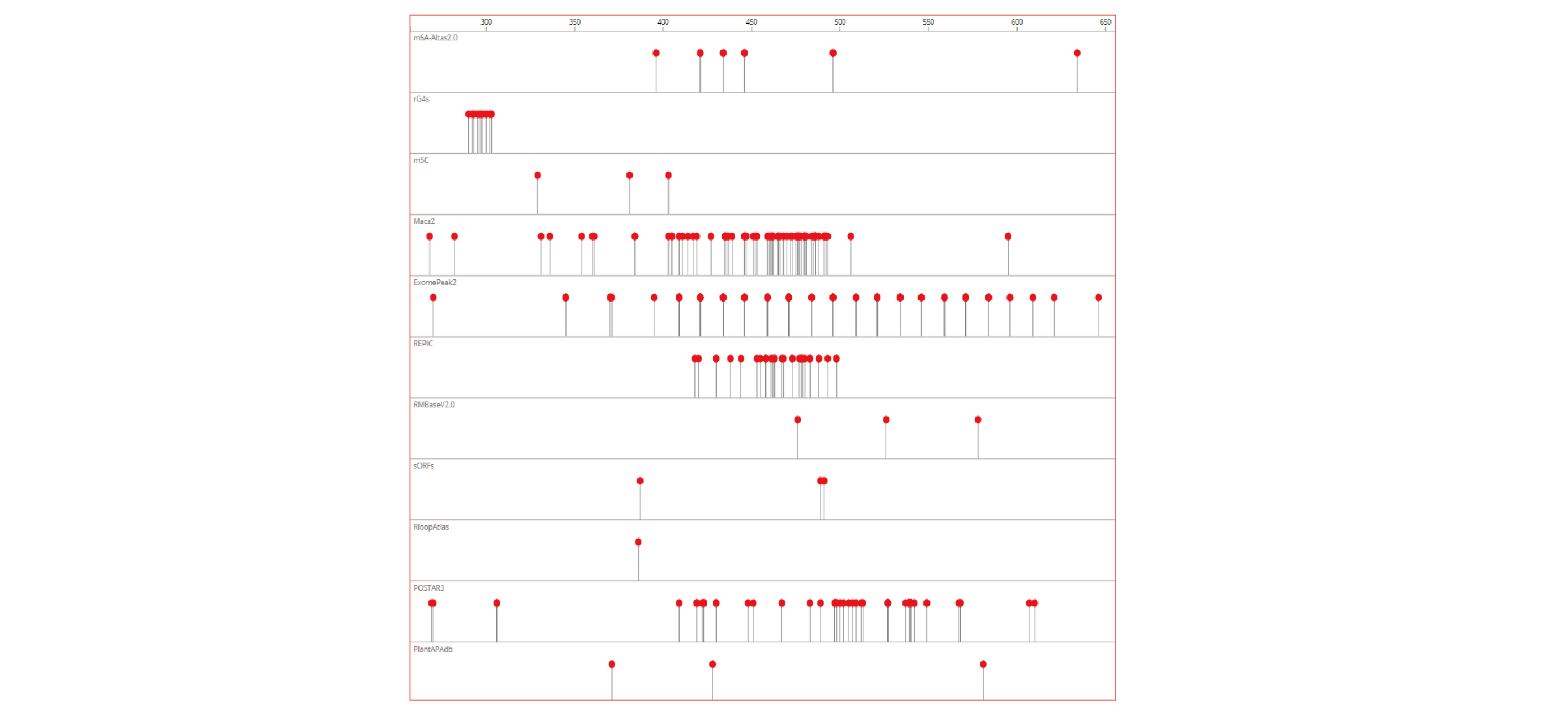
- The density/histogram map of RNA modifications to RNA exon splice junction, transcription start/end site and translation start/stop codons
- Motif identification for RNA modification sites
- Heat-map of site distribution among transcription start/end sites and translation start/end sites
- The gene functional annotation of mRNA modification sites
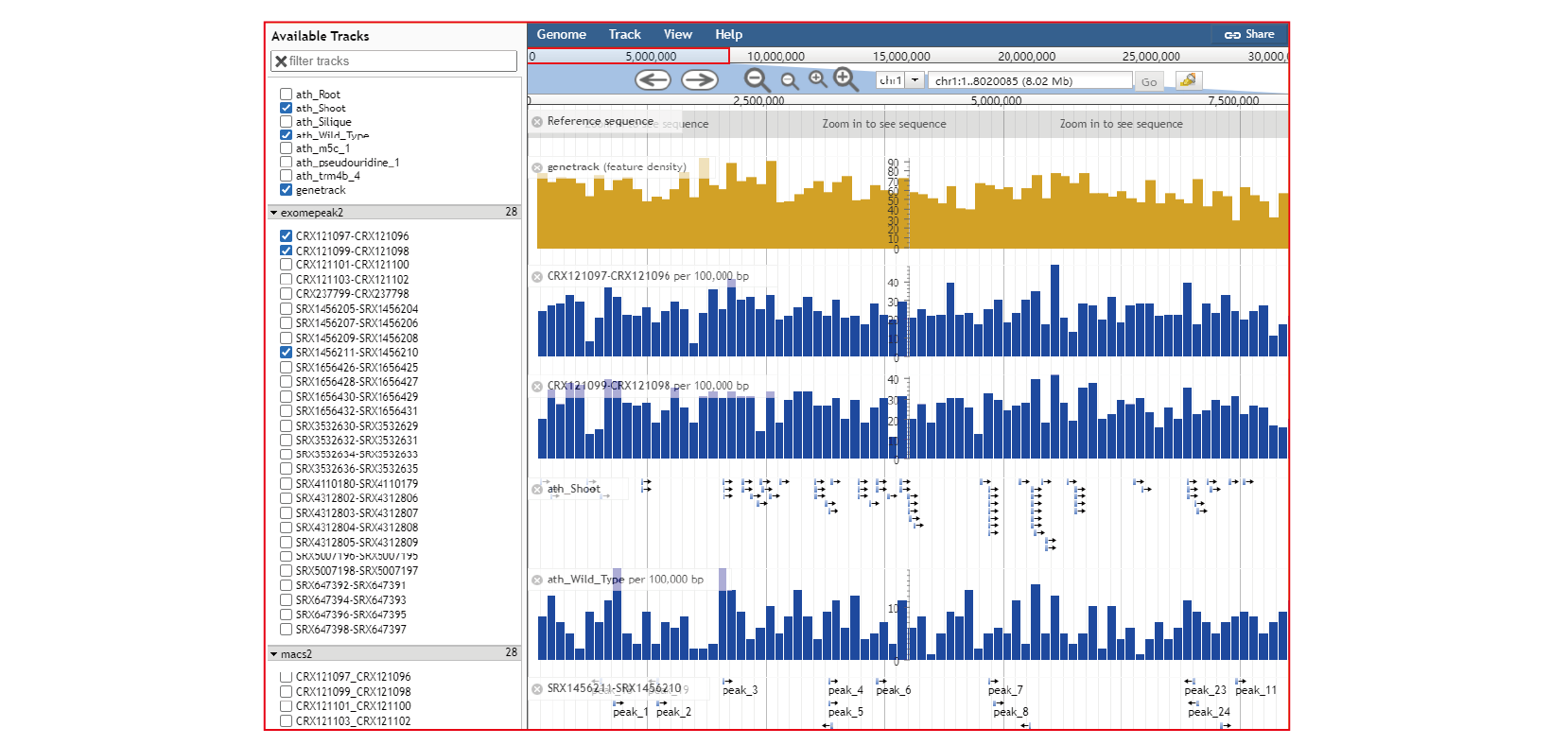
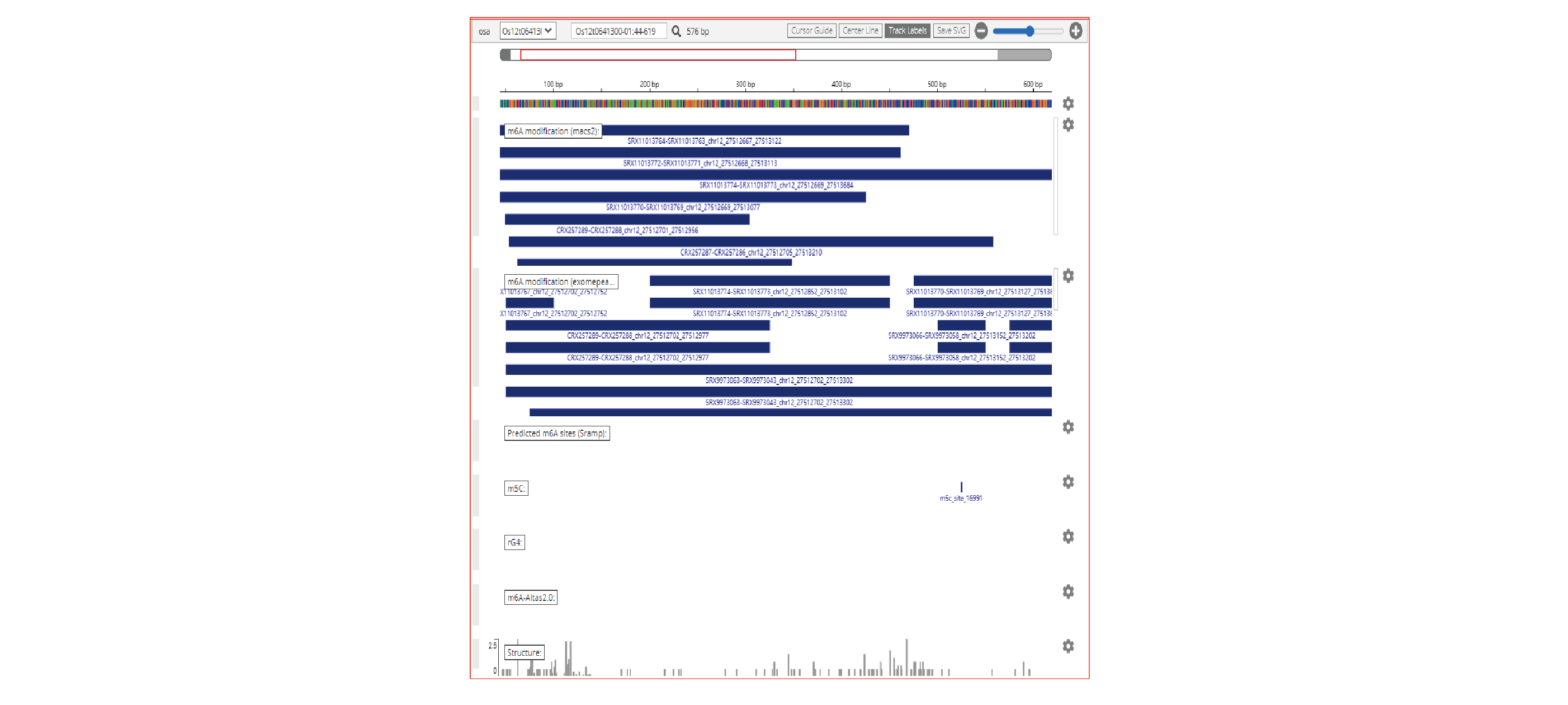
- JBrowse, mRNAbrowse,mRNAlollipop and Integrative Genomics Viewer are integrated to visualize the RNA modifications
- The RMlevelDiff tool to identify differential mRNA modifications
- The RMpantVar tool to detect mutation effects on mRNA modifications
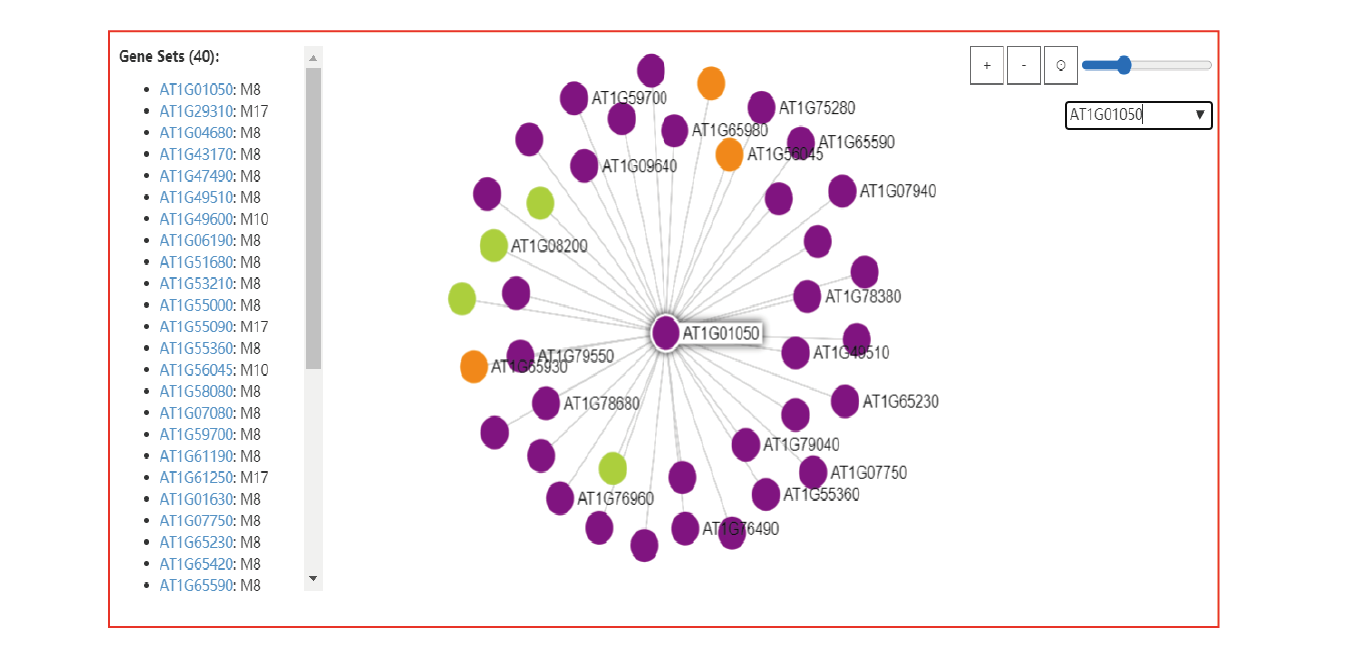
- The Netviewer tool for gene-co-methylation network analysis
PRMD: an integrated database for plant RNA modifications
Please cite us with this Reference
Recent news
Ac4C is contained in PRMD.
Blast is integrated in PRMD.
RNAmodNet is developed in PRMD.
More species are included in PRMD.
RMlevelDiff is developed in PRMD.
RMplantVar is provided in PRMD.
mRNAbrowse is integrated in PRMD.
RPMD is released.