Welcome to RNAmod (Go to new RNAmod)
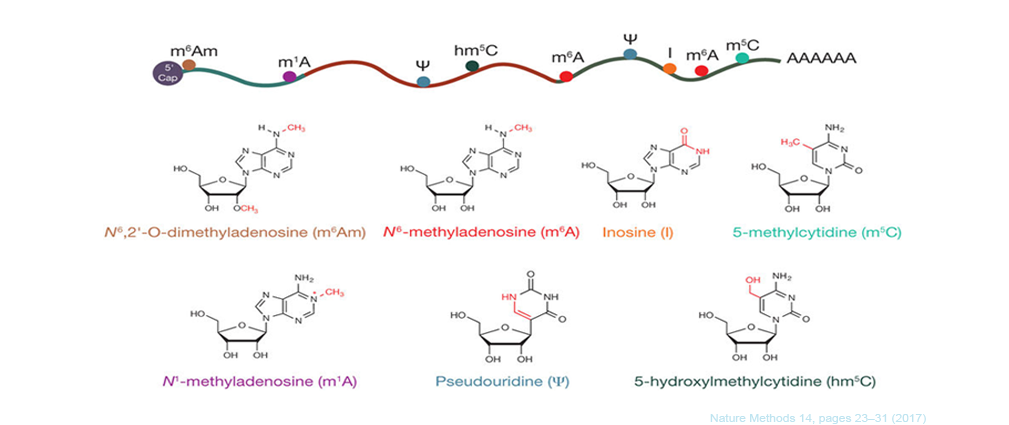
The dynamic, reversible mRNA modifications play critical roles in regulating mRNA splicing, export, stability and translation. Accordingly, mis-regulations of mRNA modifications result in multi-tissue developmental defects and cancer progression. Modifications on mRNAs are not randomly distributed, they occur on specific sequence motifs at certain locations on mRNA. For example, N6-Methyladenosine (m6A), the most abundant modification on mRNAs, is mainly located near translation start sites and translation stop sites; while N1-methyladenosine (m1A) is highly enriched at the 5'UTR of mRNA. There are more and more novel modification sites have been identified on mRNA, such as Cytidine N4-acetylation and internal m7G modifications. The localization of mRNA modification correlates with its molecular function, therefore, determining the distribution of modification across the mRNA and annotating the modified gene targets is essential for studying the function of mRNA modifications. Here we developed RNAmod, a very convenient web-based platform for the meta-analysis and functional annotation of modifications on mRNAs. RNAmod uses the commonly used BED format chromosomal location information of mRNA modifications as input, which can be generated with common peak-calling tools, such as MACS, Exomepeaks and MetaTeak, or can be easily converted from other text formats.
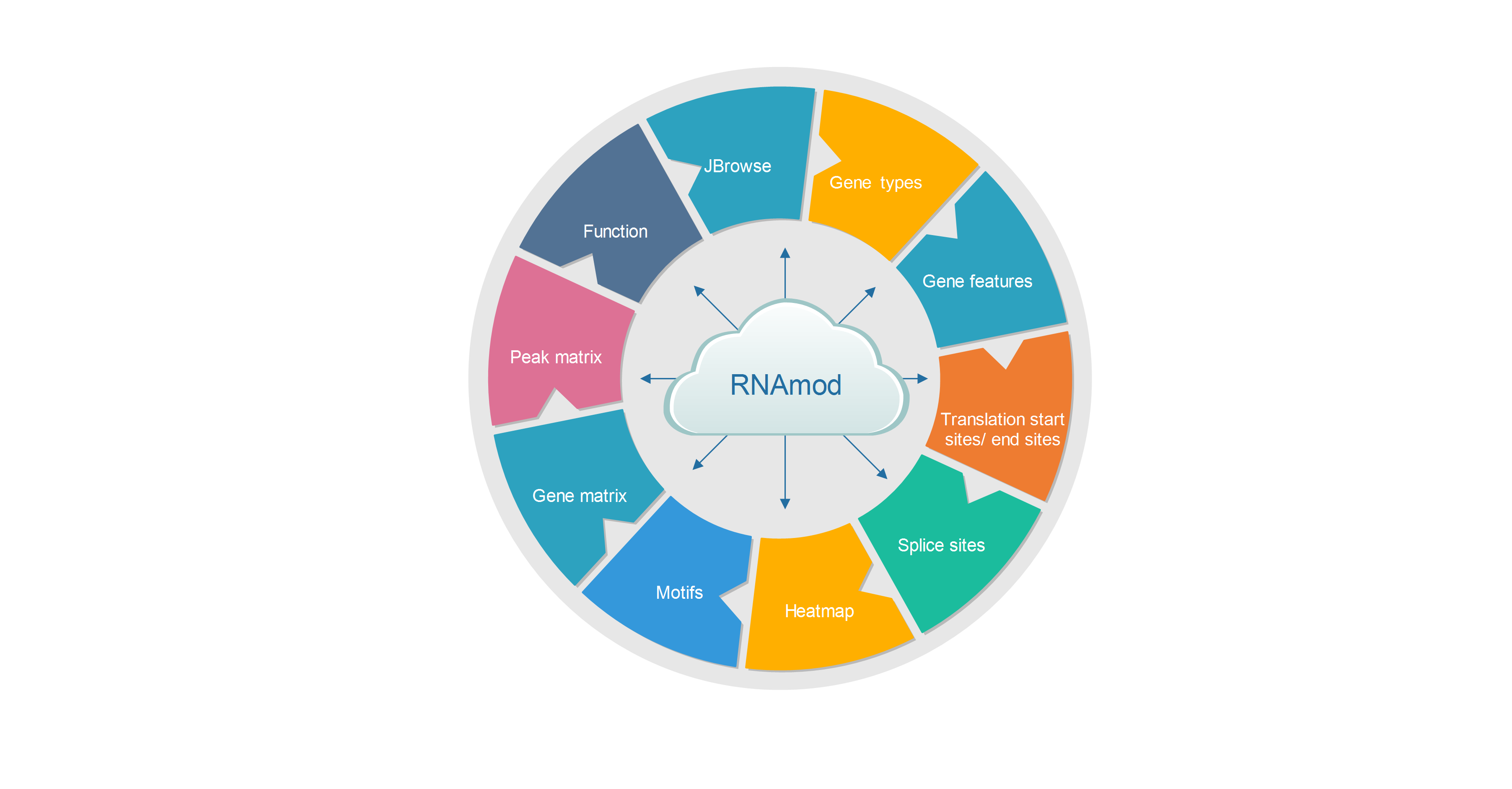
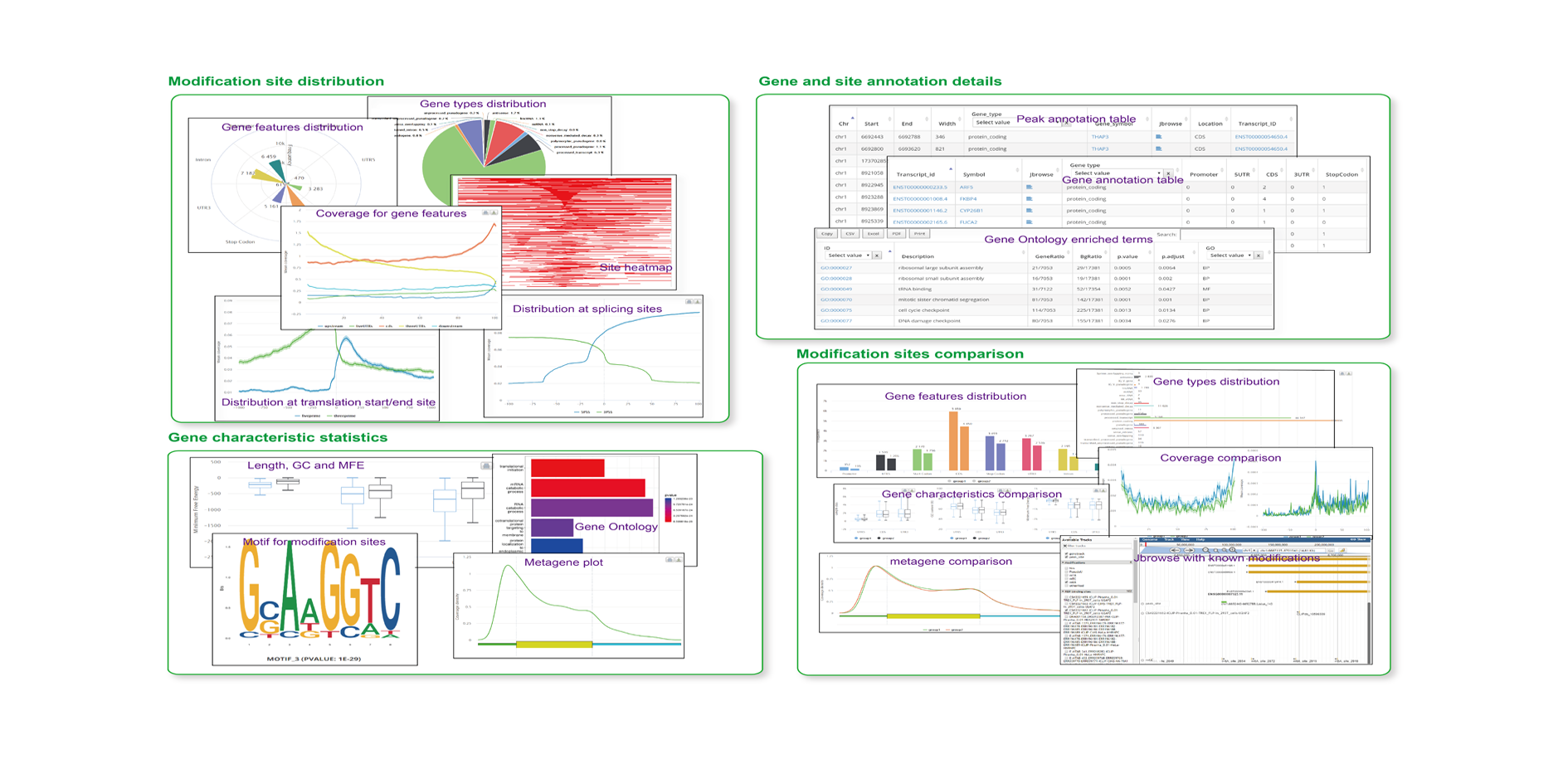
The output of RNAmod includes:
- The overall statistic is of modifications across different RNA features
- The overall statistics of modifications across different RNA biotypes
- The density/histogram map of RNA modifications to mRNA exon splice junction
- The density/histogram map of RNA modifications to mRNA transcription start/end site
- The density/histogram map of RNA modifications to mRNA translation start and stop codons
- Motif identification for RNA modification sites
- Heat-map of site distribution among transcription start/end sites and translation start/end sites
- The gene functional annotation of mRNA modification sites
- JBrowse is integrated to visualize the RNA modifications in the context of known RNA modifications and binding sites of more than 500 RBPs
- Users can conveniently study the RNA modifications sites in specific gene context
RNAmod: an integrated system for the annotation of mRNA modifications
Please cite us with this Reference
Start analysis
Help documents
Recent news
Add a new tool for prediction of mRNA m6A sites.
Large scare human and mouse tissue m6A modifications are incorporated in JBrowse (Jun'e at al, 2019, Molecular Cell; Shan et al, 2019, Nature Cell Biology).
KEGG, Reactome and Disease gene sets are added in modified gene functional annotation.
The gene case analysis is developed to allow users to analysis the modifications in specific gene context
Jbrowse is integrated in RNAmod to help users compare the modifications with known modifications and RBP binding sites
The group case functionality is developed in RNAmod
RNAmod is released to help annotating RNA modifications in 21 species.